A joint team led by Prof. Wei Xie at Tsinghua University, Prof. Zi-Jiang Chen and Prof. Han Zhao from Shandong University, has charted the translational landscapes during the human oocyte-to-embryo transition (OET). This dataset further identified a group of homeobox TFs, including TPRXL, TPRX1, and TPRX2, which are highly translated around ZGA, and are required for proper ZGA and preimplantation development. Their findings, published in Science on Sept 8, 2022, not only revealed the conservation and divergence of translational regulation during OET, but also identified critical transcription factor (TF) regulators of human zygotic genome activation (ZGA).
During the mammalian oocyte-to-embryo transition (OET), translation plays a critical role in regulating meiotic resumption, zygotic genome activation (ZGA), and early embryonic development. ZGA marks the first transcription event in a new life and the onset of the embryonic program. However, how mammalian ZGA is initiated remains poorly understood. For example, while key ZGA transcription factors (TFs) have been well characterized in other species such as zebrafish and fly, which TFs control human ZGA remains elusive. By combining the ultrasensitive Ribo-lite (Ligation-free, ultra-low-InpuT and Enhanced Ribo-seq) recently developed by Wei Xie’s group with Smart-seq2, Ribo-RNA-lite (or R2-lite) was adapted to co-profile the translatome and transcriptome across 8 stages of human oocytes and early embryos. Through comparison with their counterparts in mouse, both conserved and widespread divergent translational activities were discovered, with the latter enriched for those functioning in epigenetic reprogramming, transposon defense, and small RNA biogenesis. Species-specific translation is in part driven by different configurations of regulatory elements such as cytoplasmic polyadenylation element (CPE) and polyadenylation signal site (PAS) in the 3’ untranslated regions (3’ UTRs).
Using the R2-lite data, a group of homeobox TFs were found to become highly translated before or during ZGA, with their motifs enriched in distal open chromatin regions (putative enhancers) near activated genes upon ZGA. These TFs include TPRXL, encoded by a CPE-containing maternal transcript subjected to translation upregulation upon meiotic resumption, and TPRX1/2, which are expressed during the early phase of ZGA (minor ZGA). The joint knockdown of TPRX1/2/L (TPRX triple KD or TKD) led to severe defects in development and ZGA. About 31% of ZGA genes, which preferentially contain PRD-like TF binding motifs at promoters and nearby putative enhancers, were downregulated in TPRX TKD embryos. These TPRX target genes include a panel of downstream TF genes such as ZSCAN4, DUXB, DUXA, NANOGNB, DPPA4, GATA6, DPRX, ARGFX, and KLF5. Finally, ectopically expressed TPRXs could bind and activate a subset of ZGA genes in human embryonic stem cells (hESCs). Thus, these data revealed the conservation and divergence of translational regulation and their underlying mechanism during OET, and identified critical transcription factor (TF) regulators of human zygotic genome activation (ZGA), TPRX1, TPRX2, and TPRXL.
Prof. Wei Xie from School of Life Science at Tsinghua University, and Prof. Zi-Jiang Chen and Prof. Han Zhao from Center for Reproductive Medicine at Shandong University are the co-corresponding authors of this work. PhD students Zhuoning Zou from CLS program at Tsinghua University, Chunxin Zhang from Center for Reproductive Medicine at Shandong University, Qiuyan Wang from School of Life Sciences at Tsinghua University, and Research Assistant Zhenzhen Hou from Center for Reproductive Medicine at Shandong University are the co-first authors of this work. This work was supported by Key laboratory of Reproductive Endocrinology of Ministry of Education, National Research Center for Assisted Reproductive Technology and Reproductive Genetics at Shandong University, and Animal Center and Biocomputing Facility at Tsinghua University. This work was funded by the National Natural Science Foundation of China, the National Key R&D Program of China, the Research Unit of Gametogenesis and Health of ART-Offspring, Chinese Academy of Medical Sciences, Innovative research team of high-level local universities in Shanghai, the Tsinghua-Peking Center for Life Sciences, and the Beijing Municipal Science and Technology Commission. Wei Xie is a recipient of an HHMI International Research Scholar award.
Article Link :https://www.science.org/doi/10.1126/science.abo7923
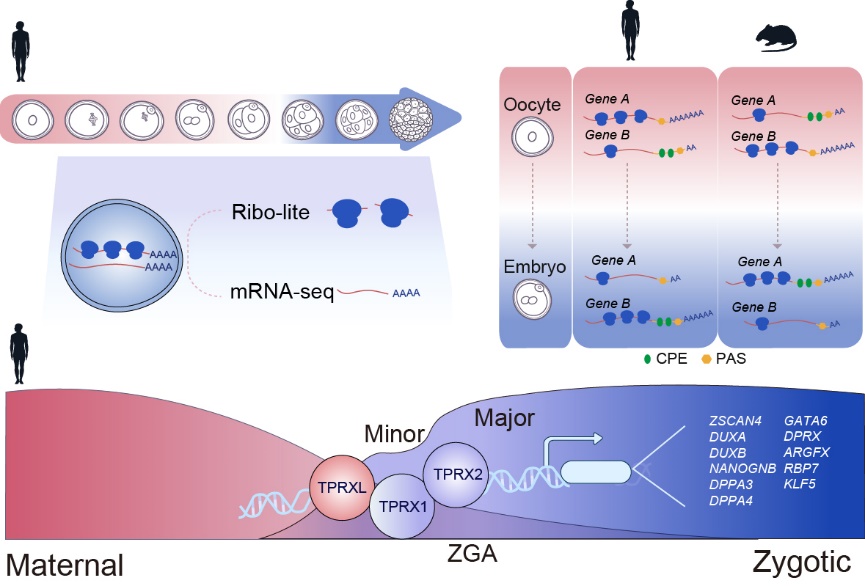
Translatome and transcriptome co-profiling in human oocytes and early embryos reveals translation regulation mechanisms and key TF regulators for human ZGA.
