Prof. Wei Xie’s group at School of Life Sciences, Tsinghua University published research in Nature on October 28, 2020, entitled “The landscape of RNA Pol II binding reveals a step-wise transition during ZGA”, revealing the activation of the zygotic genome by RNA polymerase II (Pol II) through stepwise transition. This study not only helps us to understand the fundamental mechanism underlying the onset of zygotic genome activation (ZGA), the first transcription event in life, but may also pave the way for future clinical research on early development-related diseases and assisted reproduction.
How zygotic genome activation (ZGA) occurs is one of the most basic and intriguing questions in developmental biology. In almost all studied species, transcription in mature oocytes is silenced. After fertilization, the genome remains silenced after a few cell cycles and is rapidly activated (for example, late 2-cell for mouse and 8-cell for human), accompanied with the degradation of maternal RNA, leading to the maternal-to-zygotic transition of transcriptome. Zygotic genome activation includes two steps, minor- and major ZGA. In mice, minor ZGA starts at around the middle / late 1-cell stage, with a small number of genes and certain classes of repeats being transcribed. Later work showed that widespread weak transcription also occurs genome wide, producing non-productive transcripts without proper poly-A tailing and splicing. Major ZGA occurs at the late 2-cell stage when a large number of genes is activated. Interestingly, similar to major ZGA, minor ZGA is also essential for embryonic development, as blocking minor ZGA leads to the failure of major ZGA and embryonic lethality. Nevertheless, due to the limited research materials for early embryos, many fundamental questions regarding ZGA remain unanswered. For example, why ZGA only initiates after a few cell cycles, but not immediately after fertilization? Why minor ZGA is important for early embryonic development? How is mammalian ZGA initiated?
To address these questions, the researchers focused on RNA polymerase II (Pol II), the central player of transcription machinery. Traditional ChIP-seq method, which often requires a large number of cells, is not suitable to study Pol II in early embryos. By developing Stacc-seq (in collaboration with Vazyme Biotech), a highly sensitive approach to detect the interaction between protein and DNA genome-wide, the researchers discovered that Pol II undergoes “loading, pre-configuration, and production” during ZGA. After fertilization, Pol II is preferentially loaded to CG-rich promoters and accessible distal regions in 1-cell embryos (“loading”), in part shaped by the inherited parental epigenome as shown for maternal DNA methylation. Interestingly, Pol II already initiates the re-location to future gene targets prior to genome activation (“pre-configuration”), before it later engages in full transcription elongation upon major ZGA (“production”). These results may explain why ZGA does not occur immediately after fertilization. Since drastic epigenetic reprogramming occurs after fertilization, the firing of Pol II needs to be synchronized with the maturation of chromatin, and ZGA presumably cannot start until the Pol II pre-configuration is completed.
So how does Pol II find its correct targets during pre-configuration? Interestingly, transiently blocking Pol II elongation during minor ZGA impairs the pre-configuration of Pol II and leads to developmental arrest, accompanied by aberrant retention of Pol II and ectopic expression of one-cell Pol II targets upon major ZGA. This finding reveals a key role of minor ZGA in the pre-configuration of transcription machinery for genome activation. While the exact underlying mechanism remains unclear, the researchers proposed that transcription factors may recruit Pol II to major ZGA sites, where Pol II or minor ZGA in turn stabilizes the transition, for example, by destabilizing nucleosomes and recruiting chromatin remodelers. In summary, this study identified key steps of Pol II chromatin engagement during mammalian ZGA, including a critical pre-configuration step, and revealed the essential roles of minor ZGA in Pol II transitions. These data define cornerstones for future investigations of mammalian ZGA and early development.
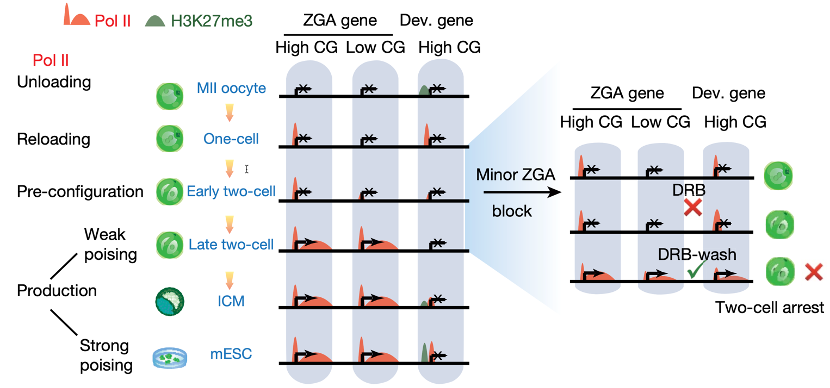
Step-wise transition of RNA polymerase II during mouse ZGA
Prof. Wei Xie from School of Life Science at Tsinghua University is the corresponding author of this work. Postdoc fellows Bofeng Liu, Qianhua Xu and Qiujun Wang from School of Life Science at Tsinghua University are the co-first authors of this work. Collaborators include Nanjing Vazyme Biotech, Prof. Jie Na ’s group from School of Medicine at Tsinghua University, and Prof. Yang Yu’s group from Institute of Biophysics at Chinese Academy of Sciences. Su Feng and Junwei Nie from Nanjing Vazyme Biotech, as well as PhD student Fangnong Lai from Prof. Wei Xie’s group, and PhD student Peizhe Wang from Prof. Jie Na ’s group also made important contributions to this work. This work was supported by the National Natural Science Foundation of China, the National Key R&D Program of China, Beijing Municipal Science & Technology Commission and the THU-PKU Center for Life Sciences. Prof. Wei Xie is also an HHMI International Research Scholar. Bofeng Liu is supported by a postdoctoral fellowship from the Tsinghua Shuimu Scholar. Qianhua Xu and Qiujun Wang are supported by postdoctoral fellowships from the Tsinghua-Peking Center for Life Sciences. The research was also supported by the gene sequencing and computing core facilities at Tsinghua University.
Paperlink: https://www.nature.com/articles/s41586-020-2847-y
