A joint team led by Prof. Wei Xie at Tsinghua University and Prof. Yi-Liang Miao at Huazhong Agricultural University has revealed how genome architecture is reprogrammed during somatic cell nuclear transfer (SCNT). Their findings, published in Molecular Cell on June 23, 2020, not only elucidated the multi-step reprogramming of 3D chromatin architecture during SCNT, but also found that an unexpected role for cohesin in repressing minor zygotic genome activation (ZGA).
Somatic cell nuclear transfer (SCNT) can reprogram a somatic nucleus to a totipotent state. Nevertheless, most SCNT embryos cannot survive to term, implying that critical epigenetic defects exist in these embryos. SCNT embryogenesis is accompanied with the dramatic chromatin reorganization induced by ooplasm. Due to the scarcity of the research samples, how chromatin is reprogrammed at the molecular level during this process, especially for the three-dimensional chromatin architecture, remains poorly understood. Using a low-input Hi-C (sisHi-C) approach, the researchers revealed the spatiotemporal dynamics of chromatin structure during SCNT. Previous work from the Xie lab and other research groups have shown that conventional chromatin structures such as topologically associating domains (TADs) are unusually relaxed in early embryos of fly, fish, mouse and human, indicating that this is an evolutionarily conserved feature. However, how and why this occurs remain a mystery. Here, researchers showed that SCNT embryos also undergoes chromatin relaxation, suggesting that ooplasm has a unique ability to convert chromatin to a relaxed state regardless of the chromatin origin. Specifically, the transferred nucleus first enters a mitotic-like state during SCNT. At the 1-cell stage, unlike fertilized embryos, in which TADs are strongly depleted, SCNT embryos show stronger TADs. TADs nevertheless become weaker at the SCNT 2-cell stage, followed by gradual consolidation. On the other hand, compartments A/B are markedly weak in 1-cell SCNT embryos and become increasingly strengthened afterward. By the 8-cell stage, somatic chromatin architecture is largely reset to embryonic patterns.
Unexpectedly, the researchers found that in both mouse embryonic stem cells and differentiated cells, removing cohesin, the key protein that mediates higher-order chromatin organization such as TADs, leads to not only disruption of TADs, but also the derepression of minor ZGA genes, including Zscan4 etc. These genes are activated prior to most of the genome during ZGA, and play important roles in embryonic development, including genome integrity and telomere lengthening. However, their activation is often defective in SCNT embryos. Finally, researchers found that pre-depleting cohesin in donor cells facilitates minor ZGA and SCNT development. In sum, these data reveal multi-step reprogramming of 3D chromatin architecture during SCNT and discovered dual roles of cohesin in TAD formation and minor ZGA repression, providing a key step towards understanding the uniquely relaxed chromatin organization in early development.
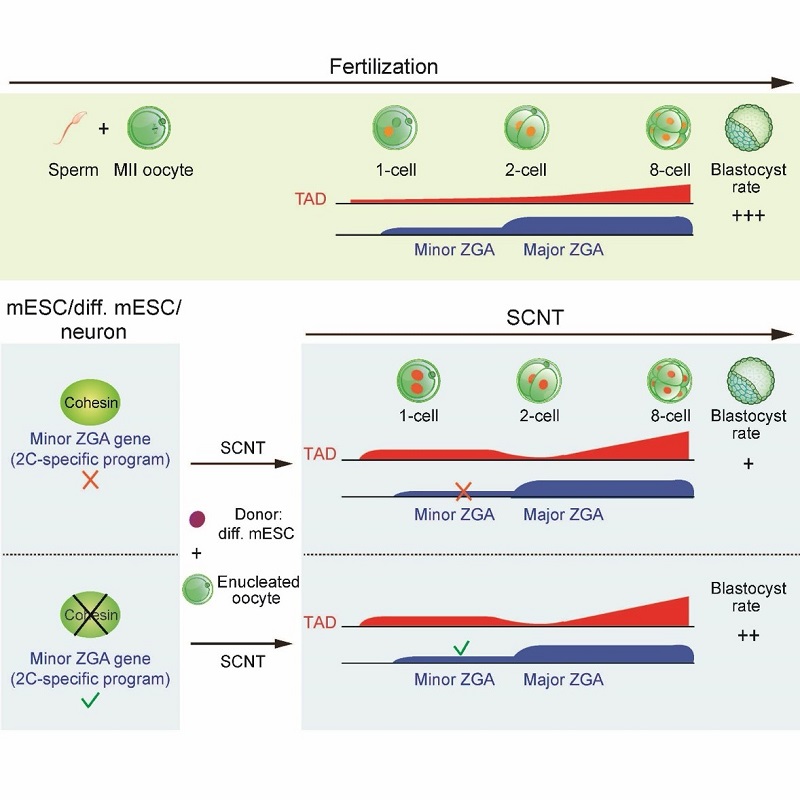
Figure 1. The schematic model for the 3D chromatin reprogramming during fertilization and SCNT, and the role of cohesin in repressing minor ZGA genes.
Prof. Wei Xie from School of Life Science at Tsinghua University and Prof. Yi-Liang Miao from College of Animal Science and Veterinary Medicine at Huazhong Agricultural University are the co-corresponding authors of this work. Ph.D candidate Ke Zhang from School of Life Sciences at Tsinghua University, Ph.D candidate Dan-Ya Wu from College of Animal Science and Veterinary Medicine at Huazhong Agricultural University, Postdoc fellow Hui Zheng from CLS program of School of Life Sciences at Tsinghua University and Ph.D candidate Yao Wang from School of Life Sciences at Tsinghua University are the co-first authors of this work. Collaborators include Prof. Kim A. Nasmyth, Prof. Robert J. Klose from Department of Biochemistry at University of Oxford and Dr. James D.P. Rhodes from MRC Laboratory of Molecular Biology at Cambridge. Qiao-Ran Sun, Xin Liu, Li-Yan Wang, Wen-Jing Xiong from College of Animal Science and Veterinary Medicine at Huazhong Agricultural University, and Qiujun Wang, Zilin Lin, Zhenhai Du, Kai Xu, Lijia Li, Guang Yu, Yao Yao, Weikun Xia, Bo Huang from School of Life Sciences at Tsinghua University also made important contributions to this study. This work was supported by the National Key R&D Program of China, the National Natural Science Foundation of China, the THU-PKU Center for Life Sciences and its postdoc fellowships, the Beijing Municipal Science & Technology Commission, competitive funding program from Beijing Advanced Innovation Center for Structural Biology, the Wellcome Trust and the European Research Council, THU-PKU Center for Life Sciences postdoctoral fellowships. Prof. Wei Xie is also an HHMI International Research Scholar.
Paper link: https://doi.org/10.1016/j.molcel.2020.06.001
